A two-year study of the new corona has been accepted for publication in an academic paper, led by COGNANO's Development Director Maeda (first author).

Recently, the Kyoto University team asked for media coverage and spread the word with "alpaca antibodies”.

We received many mentions in the web news media.
- Alpaca antibodies effective against all mutant strains of corona, Kyoto University and other research teams announce — Livedoor News
- Alpaca antibodies effective against all mutant strains of new corona, Kyoto University and others to commercialize in 2 years — The Sankei News
- Alpaca antibodies neutralize all new coronaviruses including Omicron strain, Kyoto University research group — Yahoo! News
A journalist asked me how we were able to produce universal antibodies in 2020 (within six months of the start of the pandemic). The simple answer is: "We were able to retrieve huge information about the immune response that occurs in the alpaca body and succeeded in optimizing it through computational processing," but it was difficult to convey this information verbally and I am not confident that it was understood. I am not sure if you understood me, because the questioner is in the medical field, and mathematics, machine learning, and the IT field are too different from each other. ...... I am not a stranger to this as I am one myself.
This blog will explain the Maeda paper. Since the pandemic, antibodies have been created all over the world, but only COGNANO has been able to respond to mutations. (Note: There have been cases in which the antibodies have been newly produced after a mutated form was found.) This indicates that we are finally entering an era in which biological phenomena are being converted into information and directly linked to products, rather than just developing substances from a test tube. One way to directly reflect biological phenomena is to extract human antibodies from corona patients, which is twinned with the COGNANO method. We hope you enjoy our previous blogs as we trace the real-life examples of new corona drug discovery!
How drugs are made
First, let me explain how drugs (candidate substances are called seeds: by the way, improved substances are called leads) are developed. Any drug works by binding to a target molecule; in the case of COVID-19, there are two strategies: either the enzymatic activity that allows the virus to survive and multiply is blocked by a low molecular weight compound, or the binding and invasion of the receptor on human cells is blocked by a larger antibody. There are two strategies.
In the case of small molecules
- A large number (in the hundreds of millions) of small molecule compounds and their derivatives (called derivatives) have been synthesized and are piled up in the warehouses of pharmaceutical companies, along with their rights.
- The efficacy of the candidate seeds is checked by injecting them into model cell cultures, etc. If a promising seed is found, the drug is then tested for efficacy.
- If promising seeds are found, they are modified into leads that are more effective and have fewer side effects.
- Although the robot can speed up the experiment, it is still difficult to blindly search for a billion substances, and information technology was expected to be used. Aiming to solve this problem, "forecasting companies" have already started up to simulate the fitting of small molecules to target molecules, a leading example being Atomwise in the Bay Area of North America.Innovative Approaches in Drug Discovery Using Machine LearningOur AI superplatform explores chemical space for novel molecules, enhancing drug discovery using machine learning to uncover unseen potential.
 https://www.atomwise.com/
https://www.atomwise.com/
- This is where DeepMind's AlphaFold v2.0 comes in. It has become useful because it can show the higher-order structure of any target molecule. However, AlphaFold v2.0 predicts known protein structures by machine learning, so there is no way to know the structure of antibodies created by random mutations, and the SPIKE variant of the new corona is almost impossible to predict (because it is random and there is little previously reported data).
For normal antibodies
- Special cells (hybridomas) that will make antibodies are created from mice, rats, and humans, and those that bind to and function with the target molecule (called clones) are searched for in cultured cells or in vitro molecular interaction checks.
- It is difficult to remodel antibody sites (paratopes) that contact target sites (epitopes).
- GSK(VIR)'s Sotrovimab, designed by reconstructing antibody genes extracted from a SARSCoV.1 patient 10 years ago, worked up to omicron BA.1. This was an excellent example of using a realistic biological phenomenon as it is.
Creation of VHH antibodies (nanobodies) from alpaca naive libraries
- About half of the VHH antibody development teams in the world create VHH antibodies from naïve libraries. Naïve libraries are a method of searching for effective antibodies from a large pool of antibody gene sequence information collected without vaccines from slaughtered camelids.
- The pool contains few antibodies that bind to targets never experienced by the organism (e.g., SPIKE protein). However, clones that exhibit slight binding activity may be found by chance with a probability of about 1 in 1 trillion to 1 in 100 trillion.
- To compensate for the low binding probability, a small portion of the SPIKE protein (RBD.), which is said to contact the human receptor molecule ACE2, is used instead of the entire SPIKE protein:
Receptor Binding
Using the entire SPIKE protein would be too noisy and difficult to experiment with. - To improve performance, we randomly insert genetic mutations by enzymes. Improvement of antibodies.
- The work is faster and cheaper because there is no animal manipulation, but the limit is to submit a few clone candidates.
- After this, the steps are the same as for small molecule cultured cell screening.
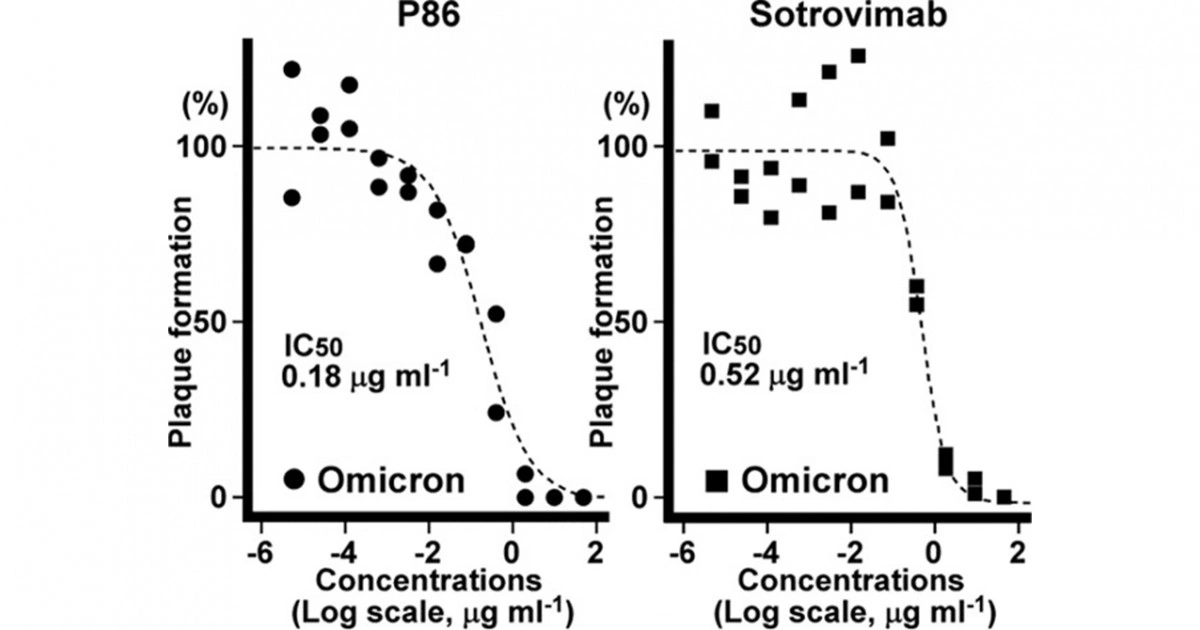
- During the pandemic, more than 10 naïve library papers were published from around the world. When development was initiated based on the Wuhan-type RBD, it was limited to about the beta type and showed characteristics of low ability to respond to mutant types. In addition, the IC50, which is an index of drug concentration, is one to two orders of magnitude inferior to the following product, which is immunized in animals.
VHH antibody production using alpaca immunization: conventional method
- Excellent VHH antibodies have been published from about 10 labs around the world using the alpaca immunization method.
- The number of antibodies (and lymphocytes that make them) targeting SPIKE increases geometrically in the body. VHH antibodies (nanobodies) are created from the collected antibody genes. Since it is important to continuously take lymphocytes, we do not kill animals.
- The principle of how organisms recombine antibody genes is known, but the rules (algorithms) for optimization are a black box. Every day, approximately one trillion (${10^{12}}$) lymphocytes recombine their genes to defend against the SPIKE protein. Theoretically, an alpaca should make more than ${10^{20}}$ attempts over a 2-month period.
- Selection based on the strength of binding to a target is called biopanning. The first step is to ensure binding. There are two methods: expression on bacteriophages and expression on yeast.
- Based on the antibody genes that pass this test, actual substances are created, and those that work as drugs are secondarily selected (screened) using 96-well cell cultures, etc. Basically, if a virus is mutated, it will be screened out from zero.
- Basically, this method is similar to the naïve library method in that if the virus mutates, you have to start over from scratch. This is because many teams choose the simple RBD as a vaccine to immunize animals or as a bait to use as a binding target, and although RBD has limitations in dealing with mutations, using SPIKE, a large protein, would be impractical because the wet selection workload would be too large. However, using SPIKE, which is a large protein, is not practical because the wet selection workload is too large.
COGNANO's Challenge
COGNANO's methodology improves the VHH conventional method, which uses only a part of the biological immune response, by making "antibody genes" informative. Since COGNANO handles enormous amount of information, it provides a bird's-eye view of the entire antibody population against the SPIKE protein, a giant natural product.
- Although we share the same approach to animal immunization, there are two fundamental differences.
- since it was known that large-scale viral mutations would occur in a pandemic, the SPIKE protein, a macromolecule, was chosen as the vaccine substance because of its extensibility to work later. 2. that we continued to immunize with mutant SPIKE every time a mutation was reported.
- IT engineers developed an algorithm to optimize the selection of binding strength and target epitopes in order to sort and organize the antibody information collected continuously over a long period of time, and to immediately determine which antibodies are useful as drugs.
- The data population of candidate antibody genes exceeds 30 million reads (counts decoded by a next-generation sequencer) after biopanning (guaranteed binding) alone.
- Of these, only information-optimized clones carefully selected by the IT team were checked in partnership with Kyoto University Hospital. (Virus experiments can only be conducted in regulated areas)
- Four of the 12 presented by COGNANO overcame the mutated form. As a result, dangerous experiments with viruses were only required for 12 clones.
- We pursued the reason why most antibodies become ineffective due to SPIKE mutation, but the antibodies created by COGNANO continue to work. To this end, we collaborated with Osaka University, JEOL, and the BINDS team to analyze the three-dimensional structure using cryo-electron microscopy, and found that the SPIKE mutation was induced in the direction of vaccine evasion, making it a weasel word. It was found to be stuck in a narrow gap.
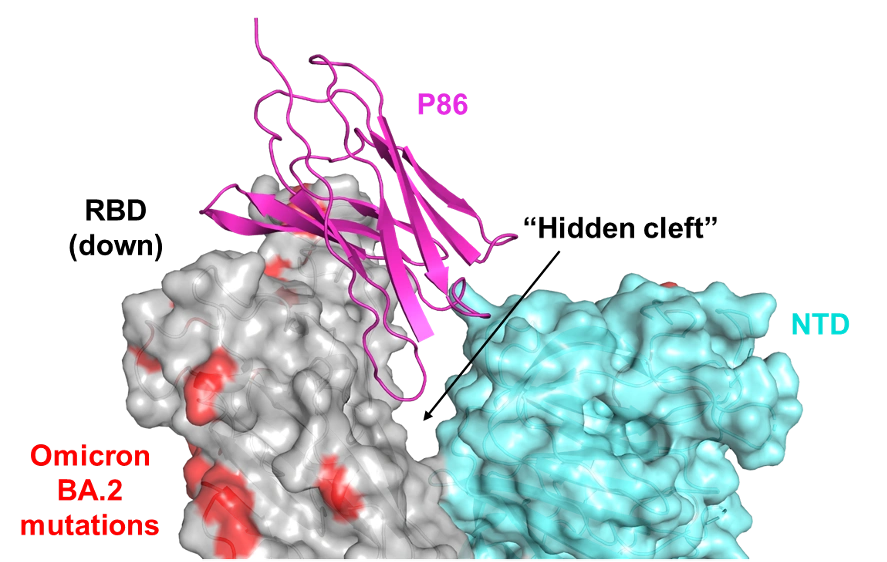
Figure description: Antibody P86, which was carefully selected by IT engineers from data of 30 million reads (counts decoded by a next-generation sequencer), binds to the SPIKE trimer by piercing the gap between adjacent RBDs. Omicron amino acid mutations are shown in red; P86 bound avoiding the red area. Human normal antibodies cannot penetrate this narrow space, indicating that the antibody binds to a "conserved structure = weak point" where the virus is not subject to evolutionary pressure. Thus, antibodies that recognize the higher-order structure of the SPIKE complex cannot be developed by a team using RBD as its material.
Methodology Summary
Reflection and Afterword
It was only when the biotechnology team and the IT engineering team worked together in a common language that we were able to achieve such results. For a team without computational power, huge information is a treasure trove, and as I wrote in my blog, I myself was lost until a few years ago. Also, what makes COGNANO different from the "structural simulations" that often appear in articles on AI drug discovery is that COGNANO starts from "evidence and labeled" real data. The alpaca mother data was actually happening in a mammalian body, not in a test tube. Nothing could be more certain.
Random and complex targets such as the SPIKE trimer mutation are difficult to track in real time with machine predictions such as AlphaFold. The immune surveillance mechanisms of living organisms are far more precise. Irreplaceable biological information can be extracted and utilized by ML and, eventually, IT technology... This is the unprecedented methodology proposed by COGNANO. In fact, a similar immune response is occurring in vaccinated people, but the human antibody gene is too complex to be easily observed. Until we decoded the alpaca gene, we had only been able to extract a small portion of nature's wisdom.
Thank you for reading this long explanation.
Now, if you have read this far, you have indigestion if I don't show you what an optimization algorithm is. The ML team is now preparing the paper, as it might be indigestion if we do not show you the Please stay tuned for this one too!