I am Hirofumi Tsuruta, in charge of research and development on machine learning at COGNANO. We have presented our work on a large-scale dataset of antigen-antibody interactions entitled "AVIDa-hIL6: A Large-Scale VHH Dataset Produced from an Immunized Alpaca for Predicting Antigen-Antibody Interactions" at NeurIPS 2023, held in New Orleans, Louisiana, United States, from December 11, 2023.
The details of the paper are explained in the blog below.

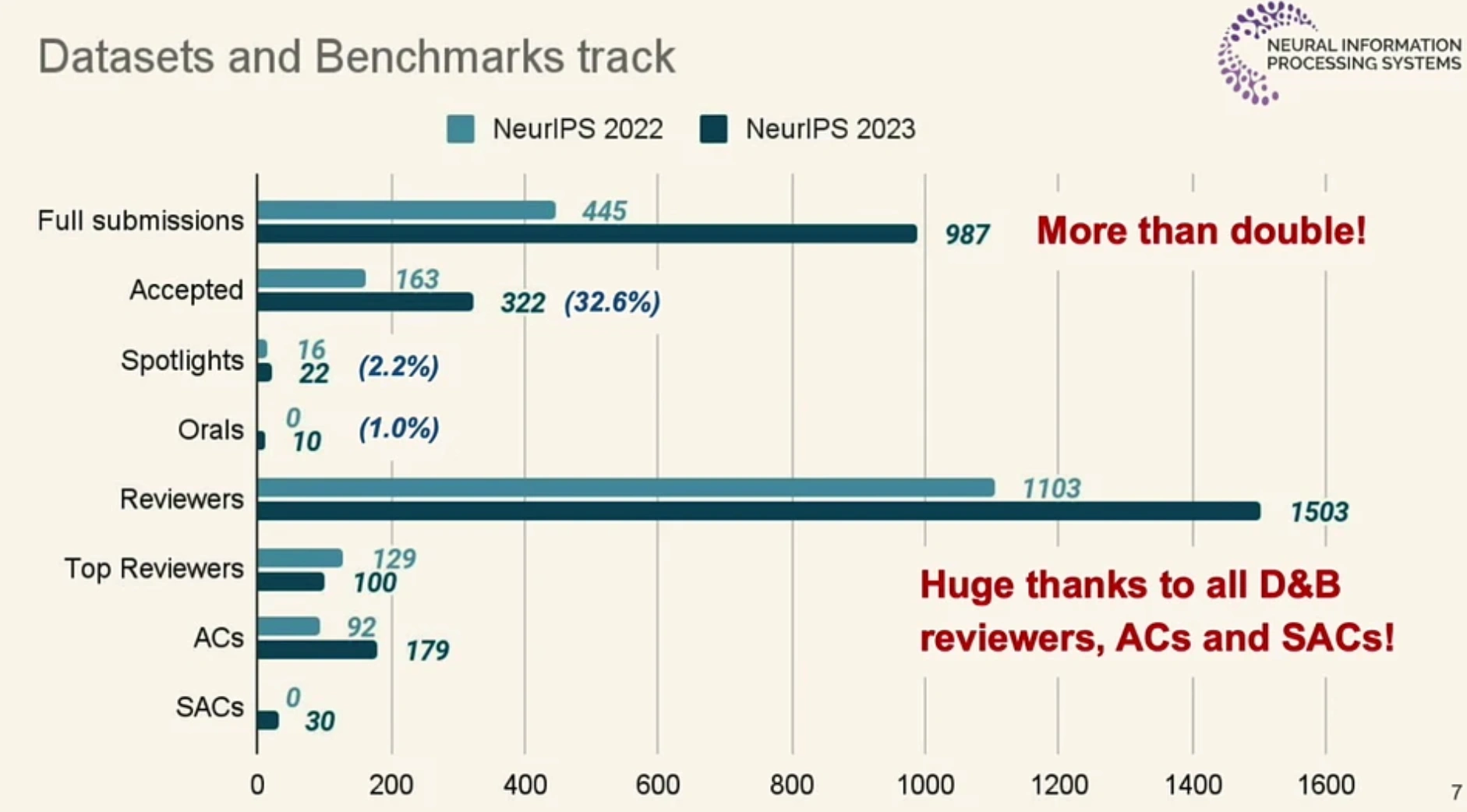
NeurIPS 2023 Datasets and Benchmarks Track
Our paper was accepted by the Datasets and Benchmarks Track at NeurIPS 2023. The Datasets and Benchmarks Track was newly established in 2021, and the number of submissions is increasing every year. This year's submissions have more than doubled compared to 2022, indicating a growing interest in data-centric research.
Our Poster Presentation
We presented our paper in the poster session on December 13th from 17:00 to 19:00 local time.

The following photos are from our poster presentation. Many people were enthusiastically interested in our presentation.



Many people came to listen to the presentations for over two hours, starting 30 minutes before the session began. During our presentation, several people were always waiting for their turn to listen to the poster presentations. Therefore, all COGNANO members, including the first author Hirofumi Tsuruta, the second author Hiroyuki Yamazaki, CEO Akihiro Imura, and CFO Yasuko Imura, explained our efforts to the people who came to hear the presentation.
Thoughts after the Presentation
Here, the authors of the paper, Hirofumi Tsuruta and Hiroyuki Yamazaki, would like to share their impressions after participating in and presenting at NeurIPS 2023.
Hirofumi Tsuruta (First author)
The first thing that surprised me when I first joined NeurIPS was the growing interest in bio-related research, such as drug discovery and proteins. There were six poster sessions in total, and every session had presentations on drug discovery and protein-related research, attracting many people. Additionally, a workshop titled "New Frontiers of AI for Drug Discovery and Development" was held this year. During our poster presentations, I always asked people who came to hear our presentations, “Are you familiar with proteins or antibodies?” and no one answered no. Several people involved in AI drug discovery at pharmaceutical companies participated in NeurIPS and came to our presentation. I was very happy and enjoyed discussions with many researchers interested in applying machine learning to drug discovery.
The presentation at NeurIPS was also a good opportunity to reaffirm the potential of our data. During the presentation, I was asked, "Do the antibodies obtained from alpacas vary with the timing after immunization or the body site from which the sample was obtained?” This is a very good question and is one of the important features of our dataset that we could not discuss in this paper. Our dataset was obtained by immunizing a single alpaca with the target molecule (antigen) four times at approximately two-week intervals and obtaining lymph node and blood samples after each immunization. We also publish metadata on when and where each sample was obtained. In this paper, we ignored this information in our data analysis, but we think it contains important information for exploring the mechanisms and dynamics of the immune system. These discussions with other researchers reminded us that our data has potential that we have yet to discuss in a single paper in NeurIPS 2023 and encouraged us to continue working hard on our research.
Hiroyuki Yamazaki(Second author)
I am Hiroyuki Yamazaki, a researcher at COGNANO. I usually work as a clinician in the Department of Hematology, and also work on data analysis, medical specimen collection, and validation experiments. I have only attended conferences in the field of medical molecular biology, and this is the first time I have had the opportunity to present at a machine learning conference. It was very exciting for me to attend a conference in a different field and at the highest international level. There was very active communication among the researchers. I was impressed by the atmosphere where I could freely exchange opinions not only with poster presenters, but also with oral presenters and eminent researchers.

As is well known, machine learning has made tremendous progress in language, image, and audio processing over the past decades. It is inevitable that the development of models in the field of medical molecular biology, perhaps one of the most complex systems, in the coming age. This is because the prediction of the structure and interaction of proteins, lipids, carbohydrates, and nucleic acids, which are the building blocks of living organisms, is directly related to the understanding of life phenomena and drug discovery. In fact, we saw a variety of approaches to drug discovery presentations at this conference. Audiences were constantly coming and going from these presentations, so we could see firsthand that this area is attracting a lot of attention.
But there's a big challenge in these areas: The amount of reliable teacher data to use as training for machine learning is very limited. As is the cornerstone of AlphaFold, there is virtually only one publicly available database, which is on the PDB website, for protein conformation prediction models. Besides, when wild-type proteins cannot be crystallized, they are often artificially mutated for conformational analysis. And the data set is not easily expandable in the near future because 3D structural analysis is quite expensive, time-consuming, and requires a high level of technical expertise. The field of medical molecular biology is much more complicated than the fields of physics and biochemistry, and the reproducibility of data is relatively low. Some multiple molecules will only interact in a normal way if they are properly integrated. Therefore, it is extremely difficult to obtain not only the presence or absence of interactions between different molecules, but also their strength as absolute objective values. However, I think there is plenty of room to develop research methods, such as building experimental systems for use in machine learning. Machine learning researchers continue to produce great theories and ideas that allow us to develop useful models with little real-world data. I am convinced that there will be more major technological innovations as the two fields literally come to understand and integrate with each other.
Future Plan
At NeurIPS 2023, we presented a data set of interactions of various antibodies with a specific target molecule (antigen) called IL-6 protein. Importantly, our data generation methodology can apply to any target molecule and will serve as a fundamental technology to build a comprehensive database of antigen-antibody interactions against a wide range of antigens in the future. In the future, we hope to publish research results on even more impactful datasets. We will continue our research with the goal of presenting our results at NeurIPS 2024 in Vancouver next year.

