Hello everyone! It’s with great pleasure that I announce today the launch of two groundbreaking projects by COGNANO: the "VHH Epitope Prediction Challenge 2026" and the "Free Access to Pancreatic Cancer Biomarker Antibodies" initiative. These initiatives represent significant milestones in our ambitious journey toward realizing "fully automated drug discovery," and are pivotal for anyone interested in the future of next-generation drug discovery technologies. Before diving into the details of those exciting projects, allow me to share our vision for automated drug discovery and its transformative potentials.
The Essence of COGNANO's Vision for "Fully Automated Drug Discovery”
Drug discovery goes beyond merely developing new therapeutic compounds; it is a comprehensive intellectual exploration that encompasses understanding the true nature of a disease and proposing therapies that addresses its root cause. COGNANO firmly believes that this exploration demands a paradigm shift in how diseases are defined – from a clinical perspective to a molecular one, and that it begins with identifying target molecules or biomarkers.
Despite being fundamental to drug discovery, current two major technologies, genomics and proteomics, have yet to identify clear targets for cancers, Alzheimer's disease and autoimmune disorders.
To tackle these challenges, COGNANO has introduced a pioneering approach called "Structure-Omics." This method views disease characteristics as molecular structure abnormalities and employs VHH antibodies as probes for comprehensive exploration. VHH antibodies, a special type of antibody derived from alpacas, excel at accurately detecting subtle structural differences. This unique capability allows them to automatically identify disease-specific structures that conventional methods have struggled to capture. The human body contains approximately ten billion unique molecular structures, known as epitopes, and antibodies are the only tools capable of distinguishing each one. Digitalizing antibodies involves translating these epitopes into an extensive volume of data. This robust, comprehensive database enables computers to identify disease-specific molecular structures with great precision. (Why VHH antibodies? See the column at the bottom of the article.)
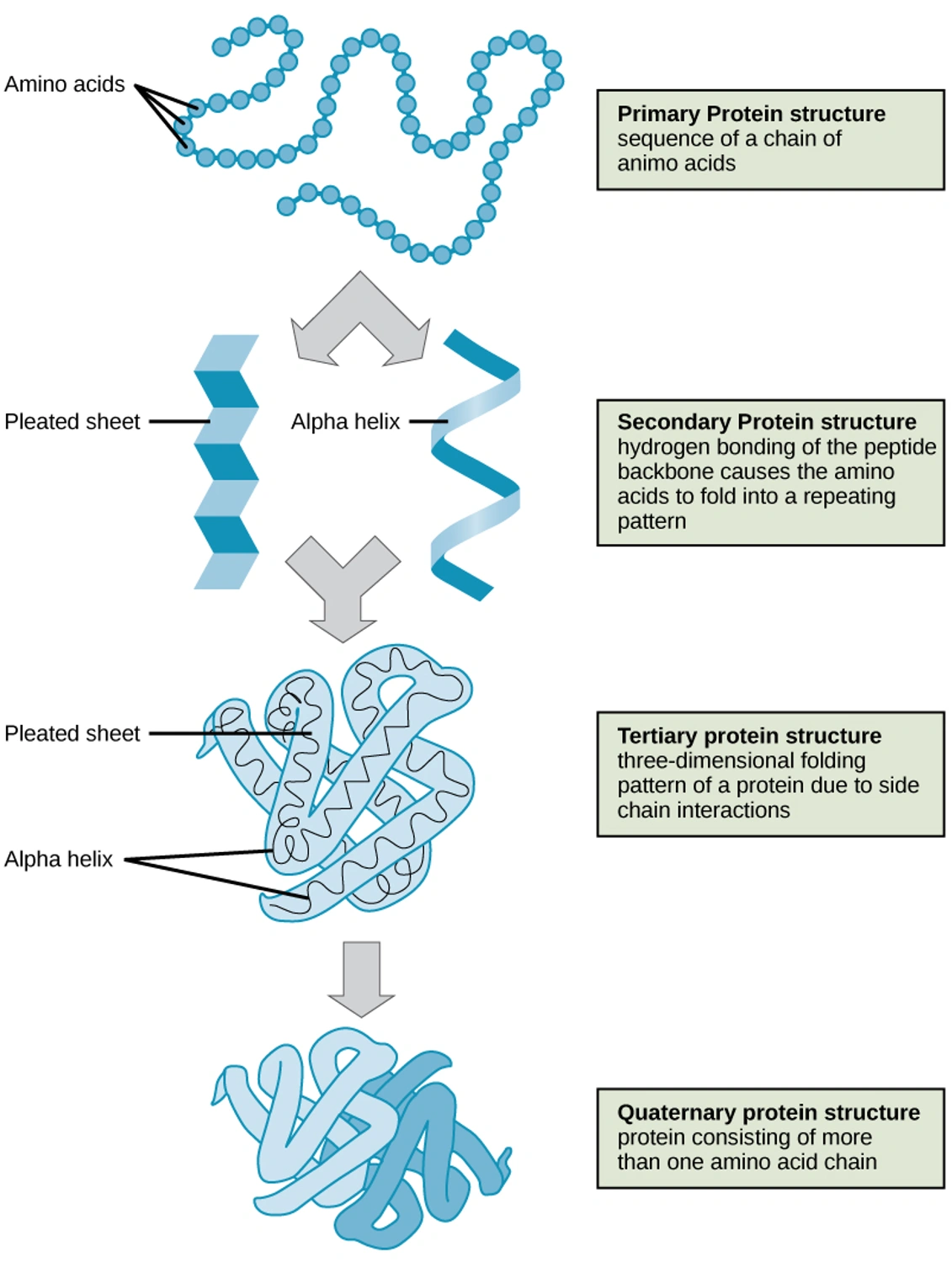
This diagram uses hemoglobin as an example to explain the principle of how amino acid chains fold to form three-dimensional structures and how molecules form complexes (the higher-order structure is explained from up to down). While general-purpose structural information is now available, there is no database for predicting disease-specific structures.
Why "automated drug discovery" has not been realized?
In recent years, the pioneering success of DeepMind's AlphaFold has marked a quantum leap in AI-based prediction of tertiary structures. AlphaFold’s breakthrough revolutionized structural biology, contributing significantly to the Nobel Prize-winning advancements in this field.
This table presents a list of currently trusted tertiary structure prediction algorithms, along with synergies and compensations in relation to COGNANO’s data. These algorithms are accessible to anyone via the internet.
Algorithm | Strenghts | Limitations | Complementarity with COGNANO Data | Recommended Use Cases |
AlphaFold (DeepMind) | High accuracy in CASP; supports complex structures | Weak in dynamics and fine epitope mapping; computationally intensive | Calibrate prediction stability with experimental labels; effective for re-ranking hit candidates | Structure identification of known targets and binding-phase screening |
RoseTTAFold (Baker Lab) | Openness, extensibility, lightweight implementation | Lower single-protein accuracy than AF; improvements depend on the community | Contributes to monitoring overfitting by antigen distribution; enables rapid validation when switching antigens | Parallel PoC with multiple antigens × multiple clones |
ESMFold (Meta FAIR) | Fast through LLM; suitable for large-scale screening | Insufficient detail in interactions; not yet adapted for function prediction | Filters generated candidates with massive “negative examples”; ensures diversity | Preliminary rough screening of de novo sequence generation |
However, despite these remarkable advancements, automated drug discovery has remained an unrealized goal. Why? The reasons are evident:
- These are all structure prediction algorithms and cannot identify disease targets.
- There is currently no existing database dedicated to the special structures that are specific to a given disease. In other words, ‘comprehensive dictionaries’ of disease information are not available.
Therefore, a team striving to develop a fully automated drug discovery system must prioritize identifying disease targets. Achieving this requires building its own new "structure databases." After identifying the targets, algorithms such as AlphaFold assist in generating new compounds effective in treating the disease.
Why can COGNANO make fully automated drug discovery reality? : Antibodies reveal a "disease map.”
Over the past decade, COGNANO has focused on developing alpaca-derived single-chain antibodies targeting various kinds of pathological cells and mutated proteins. We have conducted comprehensive screenings of VHH antibodies to identify microstructures unique to pathological cells and clarify them as biomarkers. We have pioneered the development of the world's first "structural biomarkers" for several challenging human cancers, including pancreatic cancer, bile duct cancer, and triple-negative breast cancer.
These antibodies not only provide critical disease insights but also serve as seeds for drug discovery, enabling rapid development of therapeutic antibodies. Additionally, they offer structural information that accelerates lead optimization and improves side effect prediction. COGNANO firmly believes these advancements mark the beginning of "fully automated drug discovery."
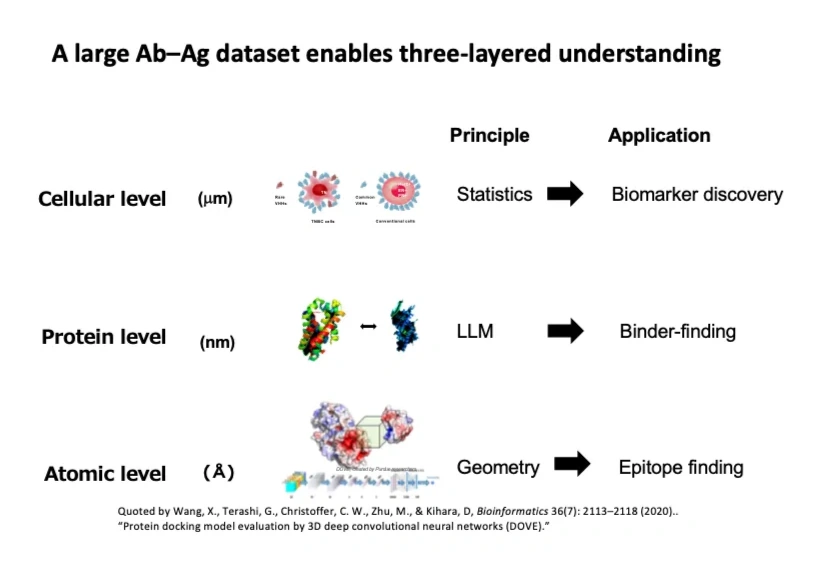
Figure illustrates the potential applications of antigen-antibody big data at three different bio-molecular levels. By analyzing antigen-antibody big data, we can distinguish differences at cellular, molecular and atomic levels, contributing to the discovery of novel biomarkers, seed design, and deeper insights into the theory of intermolecular interactions. Since antibodies are represented by sequences of amino acid alphabets, large language models can be applied effectively at all levels.
Quest for the World to Overcome the Final Challenge: Epitope Prediction
Global Call: Take on the 2026 VHH Epitope Prediction Challenge! The key obstacle to achieving fully automated drug discovery lies in the absence of technology capable of accurately predicting the epitope that an antibody binds to, solely based on protein sequences. Developing such technology could reduce the drug discovery timeline by more than a year.
To inspire AI researchers worldwide to address this demanding challenge, COGNANO is launching the "VHH Epitope Prediction Challenge" in 2026. The focus is on six lines of antibody sequences targeting Her2 molecule. Participants are tasked with predicting binding sites based on sequence data and competing for accuracy. They are welcome to use any approach, whether using existing algorithms like AlphaFold or their own models. Outstanding contestants will be rewarded with cash prizes, and celebrated for their contribution to the global drug discovery community.
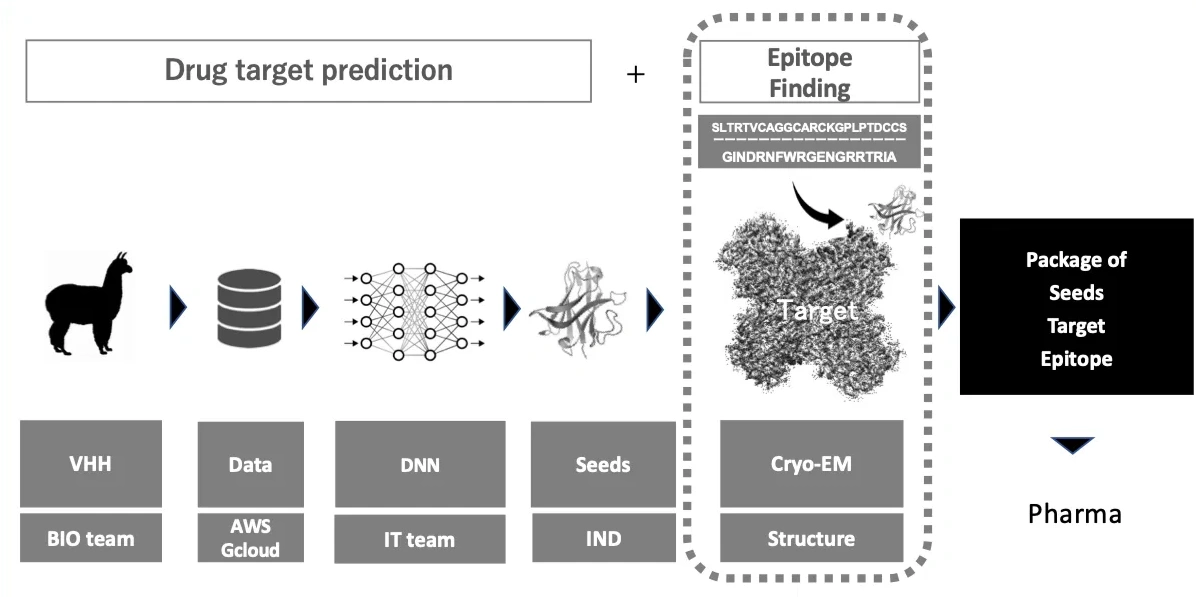
This figure illustrates the workflow of the COGNANO platform, designed to identify novel target molecules from big data on alpaca single-domain antibodies and develop optimal antibodies as seeds. The VHH Epitope Prediction Challenge in 2026 invites AI researchers to address the traditionally academic task of "epitope and three-dimensional structure identification," as indicated by dotted circles.
Free Access to Pancreatic Cancer-Specific Antibody Panels
COGNANO is launching a global program to provide established biomarker antibodies specific to pancreatic cancer, free of charge, to researchers worldwide. The initiative addresses the pressing clinical challenge of the lack of clear biomarkers for pancreatic cancer. In collaboration with public research institutions, we offer these antibodies on a non-profit basis, and we also invite interested parties to share their data and research findings to foster collaborative progress.
New World with Automated Drug Discovery on the Horizon
The inability to identify disease-specific targets is a significant obstacle to achieving fully automated drug discovery. What is often referred to as ‘Unmet Medical Needs’ largely stems from this challenge. However, we have reached a stage where, once a disease-specific target is identified, algorithms like AlphaFold can assist in developing potentially curative agents. COGNANO is tackling this immense challenge of disease-specific target identification by harnessing the natural wisdom of the body’s immune system - VHH antibodies. By comprehensively screening and converting the immune system’s natural processes into digital information, we can uncover disease targets, whether they are genetic anomalies, mutations in amino acid sequences or molecular structural differences. Identifying them brings us one step closer to our dream, fully automated drug discovery. As a small Kyoto-based venture, we are steadily building a groundbreaking "disease-specific molecular structure database," an unprecedented source in medical science.
For Al researchers, medical professionals and investors
We invite Al researchers, medical professionals and investors who share our passion to join us in this endeavor. It’s our great honor to pioneer this medical breakthrough, working hand in hand with you.

COGNANO, Inc. https://cognanous.com/contact
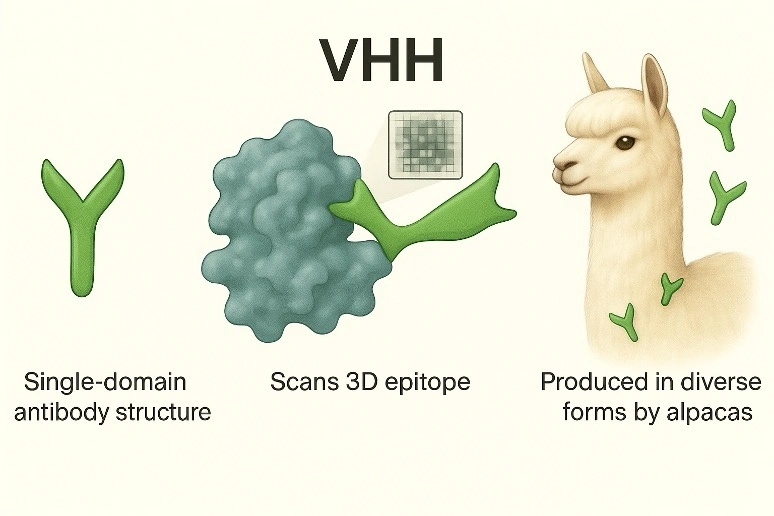
1. Simple Structure Enables Efficient Data Analysis
Antibodies from camelids (such as alpacas) consist of only heavy chains. This simplifies antibody gene analysis and significantly reduces the cost of sequencing using next-generation sequencers (approximately one millionth of conventional antibody). The resulting amino acid sequences can be used like barcodes and applied to deep learning-based data analysis.
2. Reaching Small Target Spaces
VHH antibodies are extremely small compared to typical antibodies, allowing them to efficiently access targets with complex structures. Because specific VHH antibodies accurately reflect the structural features of target molecules, they can be used as 3D pixels to scan targets.
3. Naturally Generated Diversity in Vivo Is the Greatest Strength
In medium-sized animals like alpacas, over one trillion lymphocytes constantly generate and evolve new antibody sequences. The enormous diversity that can only be reproduced in vivo and the ability to select superior antibodies are the greatest value of VHH antibodies. Real-life data obtained without causing pain to animals guarantees a quality and quantity that cannot be reproduced by in vitro procedures.

